Welcome:
The UMD software was developed not only to create various locus-specific mutation databases (LSDBs) but also numerous analyzing tools. It was thus critical to optimize the database structure and use an appropriate programming tool to meet these two goals. Among the various languages available, we chose the 4th Dimension® package from 4D because it had many advantages: 1) it runs on both Macintosh and PC platforms, 2) it allows the creation of relational databases, 3) it has a complete language of more than 700 commands, 4) it has a graphic interface and can create dynamic HTML pages, giving easy access to direct on-line queries via the Web, and 5) it has a compiler to create optimized software.
Database structure
To avoid the many errors found in publications of mutations (up to 10%) which include wrong nucleotide or AA position, reference to a wrong sequence or misinterpretation of the mutation, we built a specific structure including two tables called “Genetic Code” and “Gene Sequence.” The “Genetic Code” table is common to all LSDBs developed with UMD and contains the human “codon usage” genetic code. For each codon the amino acid (AA) three letter code and the amino acid mutability value are available. The “Gene Sequence” is specific for each LSDB. It includes for each AA position the wild-type codon and phylogenic data, defined by curators using a numeric value (conserved AA among mammals (value = 2), vertebrates (value = 3) ...). The use of these two tables secures the data entry, avoiding typing or numbering errors. Concurrently, a “Mutation” and a “Clinical Data” table were created. The “Mutation” table is the central part of the structure. It is linked to all other tables. It includes many data, with minor differences from one LSDB to another.
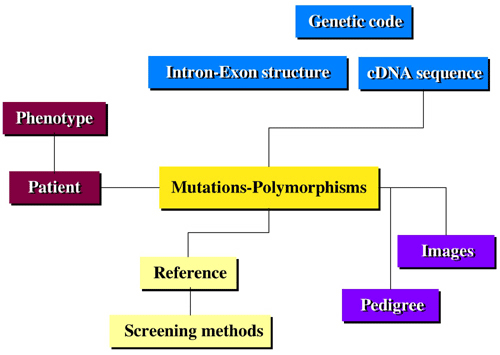
To reduce typing errors and facilitate the input of data, the software automatically checks and calculates various data, such as wild-type codon and AA, mutant AA, exon number, mutational event, mutation type, involvement of a CpG or a pyrimidine doublet, localization of the mutation in a highly conserved domain and/or in a structural domain and modification of the restriction map. Finally, when the mutation is a deletion or an insertion, the UMD software searches automatically for the involvement of a repeated sequence that could account for the mutational event. In addition a routine automatically names the mutation according to the international nomenclature. Use of the 4D SGDB gives access to optimized multicriteria research and sorting tools to select records from any field. Moreover, several routines were specifically developed.